Tutorials Overview
We provide video tutorials and links to resources in three areas:
-
Patients’ therapeutic odysseys & challenge overview. Understand the clinical motivation, challenge structure, phase timelines, expected inputs/outputs, question types, and rationale for
evidencefields. -
Biomedical resources for evidence. Review current clinical workflows for treatment discovery, with references to key datasets and resources for programmatic retrieval by models.
-
Compute environments & model access. Learn about Google Cloud Platform (GCP) environments, using Hugging Face for models and data, and core concepts like Retrieval-Augmented Generation (RAG).
Therapeutic Identification Workflows
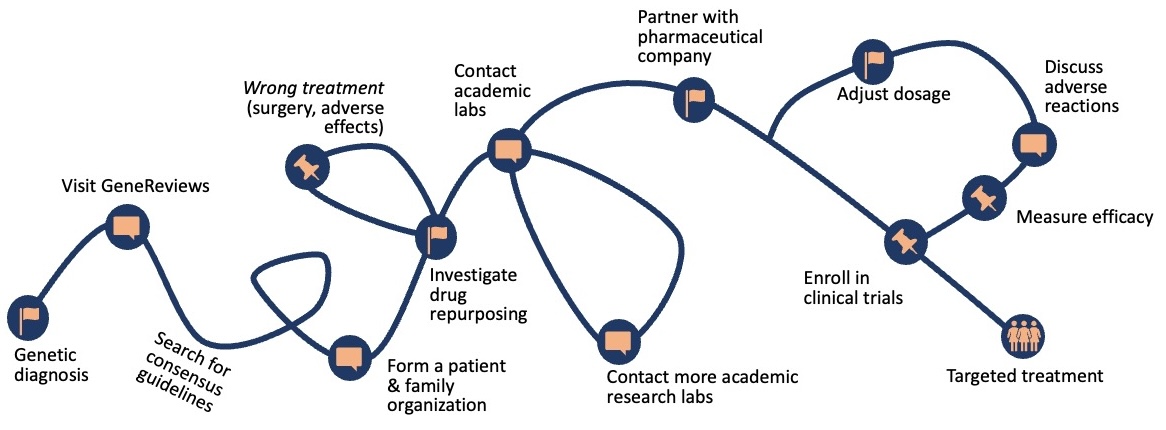 Finding treatments for patients with rare genetic disease diagnoses is a long, frustrating, largely manual process. Targeted treatments exist for only ~5% of rare disorders (Kaufmann et al. 2018).
Finding treatments for patients with rare genetic disease diagnoses is a long, frustrating, largely manual process. Targeted treatments exist for only ~5% of rare disorders (Kaufmann et al. 2018).
Biomedical Resources
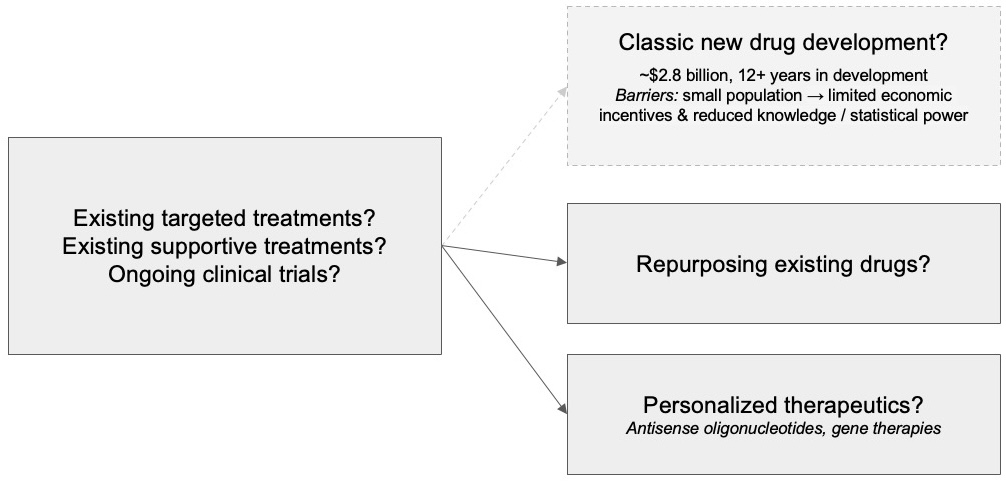
Once a clinician receives a genetic testing report with a confirmed diagnostic variant, the immediate next steps are to identify whether there are:
- existing targeted therapies, FDA-approved treatments for this specific genetic diagnosis
- existing supportive therapies, FDA-approved drugs with other primary indications but for which there is peer-reviewed literature suggesting its efficacy for this specific genetic diagnosis
- ongoing clinical trials for this specific genetic diagnosis where the patient meets all inclusion criteria and no exclusion criteria
- drugs that can be repurposed for off-label usage based on experimental evidence and/or molecular functionality rationale
- opportunities for personalized therapeutics such as antisense oligonucleotides or gene therapies based on the genetic variant location, impact, and affected tissue(s)
Learn more about resources used to manually identify therapeutic avenues:
Biomedical Resources (Nonexhaustive)
- Established (and in trial) Targeted and Supportive Therapies
- PubMed – A searchable database of biomedical literature, including research articles relevant to genetics and molecular biology.
- GeneReviews – Expert-authored, peer-reviewed disease overviews focused on the diagnosis, management, and genetic counseling of inherited disorders.
- ClinicalTrials.gov – A registry of clinical trials, providing information on ongoing or completed studies involving genetic conditions and therapies.
- Druggability Information
- ClinGen Dosage Sensitivity – Curated data on gene dosage sensitivity (haploinsufficiency and triplosensitivity) and gene-disease validity classifications.
- PHAROS – An interface to the Target Central Resource Database (TCRD) providing information on the druggability and function of human proteins.
- DrugBank – A richly annotated database of drugs and drug targets, including FDA-approved therapies and molecular interactions.
- Variant Amenability to Personalized Therapies
- N=1 Collaborative (“N1C”) - Initiative to clarify and streamline development of individualized medicines, centralizing knowledge, data, and safety information.
- Gene-Phenotype Associations
- ClinVar – A public archive of interpretations of clinically relevant variants and their supporting evidence.
- OMIM – Online Mendelian Inheritance in Man; a comprehensive catalog of human genes and genetic disorders with phenotype descriptions and molecular relationships.
- Geno2MP – A resource linking rare variants in exomes to de-identified phenotypic profiles, used for rare disease discovery and matchmaking.
- DECIPHER – A database of chromosomal imbalance and pathogenic sequence variants with phenotype correlations and genome browser integration.
- NCBI Gene – Comprehensive gene-specific information including nomenclature, sequences, pathways, and bibliographic links.
- GenCC – The Gene Curation Coalition; a unified platform of gene-disease validity assertions from multiple expert groups.
- Variant, Gene and Pathway Information
- Ensembl – Genome browser with rich annotation of genes, variants, comparative genomics, and regulation for multiple species.
- UniProt – A comprehensive resource for protein sequence and functional information, including isoforms and domain annotations.
- Mutalyzer / VariantValidator – Tools for validating and correcting variant nomenclature according to HGVS standards.
- gnomAD – Aggregated population allele frequency data for assessing variant rarity and constraint metrics across diverse ancestries.
- GTEx – The Genotype-Tissue Expression project; provides data on tissue-specific gene expression and eQTLs across human tissues.
- VastDB / ExonSkipDB / Snaptron – Databases and tools for exploring alternative splicing events and transcript usage from RNA-seq data.
- Resource aggregation platforms
- MobiDetails – A meta-resource that aggregates variant-level information and prioritization tools for clinical variant interpretation.
- Franklin by Genoox – A clinical-grade platform that integrates multiple databases and tools for variant classification and evidence curation.
Computational Resources
Hugging Face is a universal hub for:
- AI models: ~1.6M models across various tasks
- Datasets: ~375K for training and evaluation
- Deployment: single-click deployment of your project (across multiple providers) using shareable self-contained “spaces”
- User Interfaces: build and share ML apps with user interfaces (UIs) using Python on Gradio
- Education: free courses and certifications
Learn more about accessing data and deploying models in Hugging Face:
Challenge-Relevant Concepts
- Retrieval Augmented Generation (RAG)
- Retrieve up-to-date, specialized, and verifiable information from trusted knowledge bases
- Insert this information into your “model context” in order to generate answer(s) using in-context information
- Example: Almanac, NEJM AI
- Model Context Protocol (MCP)
- Standardized protocol to allow LLMs (from any provider) to use context (i.e., as required for RAG)
- Building MCP Servers with Gradio
- Example ClinicalTrials.gov MCP server on Hugging Face: Embeddings of “brief description” column from entire Clinical Trials database
- Example PubMed Medline MCP server & Gradio demo on Hugging Face: Embeddings of the description/abstract of every article on PubMed
- AI Agents
- MCP Client and a “While Loop” to simulate a chatbot
- Example AI-Tx Challenge AI Agent Demo on Hugging Face
- Constrained Generation
- LLMs are bad at generating valid JSON (required per our Challenge Output Requirements)
- Open libraries include “Outlines” for context-free grammars (CFG) and JSONFormer



